Protein Folding Problem, Origin of Life and Australia
Basic building block of life: Proteins But Life/Nonlife Line is Blurred
Protein Folding
Proteins are more than meat. Proteins are made up of 20 amino acids in the human body. [In nature, there are 500 amino acids].
You can visualize amino acids as beads in a chain: Each bead is an amino acid and the protein is the chain. All the proteins fold in a certain way to perform a specific function. This is the key to structural biology. If a protein folds wrongly, it can cause a disease: (Type 2) diabetes, Alzheimer's, Parkinson's, ALS (what Stephen Hawking had) are common examples.
Cyrus Levinthal estimated in 1969 that large proteins can be folded in 10^300 ways. Even a small one can be folded in 10^50 ways. If a protein sequentially tries out all possible shapes to get to the right shape at the rate of 1/1000 per second, it would take more time than the existence of the universe. Yet, most proteins take micro or milliseconds to correctly assume the right fold. It proves that protein folding is not a random walk in 3D.
Computational problem: If we know the amino acid sequence, can we predict what produces the most stable shape of the protein that the sequence of amino acids produce? To be sure, there can be more than one stable sequence. Prion (the badly folded protein that causes the Mad Cow Disease) is an example of a multiple possible stable sequence.
Bottom up approach - observe a large number of protein folds and find a regularity that will predict correct folds with accuracy. For that, you need a large number of proteins with known structures. Advances in cryogenic electron microscopy (Cryo-EM) have produced a large number of protein fold pictures in 3D in nature.
2017 Nobel in Chemistry was awarded for this discovery.
The remarkable fact: none of the three winners are chemists.
Jacques Dubochet, Joachim Frank are biophysicists, and Richard Henderson is a biologist.
To find how well your model fits the real proteins, you need a measure to quantify the error. The standard Root Mean Square Deviation (RMSD) turns out to be inadequate for quantifying errors in protein models. Adam Zemla, a Pole with a PhD in computational mathematics from Moscow University, devised a test called the Global Distance Test (GDT). It assesses the similarity between two structures by comparing the distances between alpha carbon atoms. The GDT has a range between 0 and 1. A 0 represents a totally dissimilar structure and 1 total similarity - the perfect score.
[Mahalanobis distance is a statistical measure used to quantify the distance between a point and a distribution, considering the covariance structure of the data. So, it is not adequate for protein folding problems.]
CASP (Critical Assessment of Protein Structure Prediction) is a long running competition among protein folding researchers. It started in 1994. The method of evaluation was not standardized. It changed with the Global Distance Test (GDT) - developed by Zemla in 2003.
State of the Art in Protein Folding
Until 2014, the best prediction got a score of 0.4 in the CASP competition. The following biennial competition saw the entrance of DeepMind. It developed a method called AlphaFold - to attack the folding problem. In 2016, it got a GDT of 0.59. In 2018, with a new version called AlphaFold 2, it got 0.89. DeepMind made their method public and retired from the competition.
What is the secret sauce? It uses an Artificial Neural Network model with several hidden layers (so called Deep Network) plus some recent AI architectural innovations. It is a diffusion model like Dall-E.
How big was the jump in modeling by DeepMind? Until 2018, there were 150,000 models of protein structures. DeepMind pushed it out to 200,000,000 in 2020 - every protein known to exist in nature and then some. In comparison, John Kendrew took 12 years to create the first protein structure for which he got the Nobel Prize in 1962.
In 2024, three researchers got the Nobel Prize in chemistry for constructing protein structures of incredible complexity. Demis Hassabis and John Jumper have developed an ANN model to solve the protein folding problem: predicting a protein's complex structure. Neither of them is a chemist.
David Baker, the third winner, got half the prize money. He has succeeded with the almost impossible feat of building entirely new kinds of proteins with desired function that does not exist from scratch using computational methods. Then, he and his lab created a synthetic DNA that encodes this protein. Put that synthetic DNA into a bacteria. The bacteria becomes the factory to produce the protein and then check if the protein does have the desired function.
Protein Folding and Origin of Life
A link between protein folding and origin of life (summarizing Kocher and Dill - Origins of Life: The Protein Folding Problem all over again? PNAS, 2024).
How did specific useful protein sequences arise from simpler molecules at the origin of life? This seemingly needle-in-a-haystack problem has remarkably close resemblance to the Protein Folding Problem. Origins of life must have come only after there was an operative evolution mechanism which selects on phenotype, not genotype. For this to work, there has to be some positive feedback mechanism.
If there is a golf course landscape, finding the hole randomly has a very low probability.
On the other hand, if the surface was undulated in the first place, the selection might lead to a local hole. Once in the local hole, it becomes easier to jump to a deeper hole - statistically speaking.
In the case of the folding problem, there are catalysts that make all possible future states not equally likely. There are hydrophobic and polar proteins. Thus, the local minima can be found fast. [Mathematically, if P≠NP, P complete problems can be tackled in polynomial time.] If local minimum is attained, by Feynman’s Brownian Ratchet mechanism, it would take much less time than Levinthal’s postulation alluded to above.
Life Began in Australia (Maybe)
Jack Hills (Western Australia) carbon evidence
The stable isotope ratio of carbon is often used as an indicator of life in old rock samples. A high carbon-12 to carbon-13 ratio suggests the carbon has been processed by living organisms, because some metabolic enzymes involved in fixing inorganic carbon ‘prefer’ carbon-12.
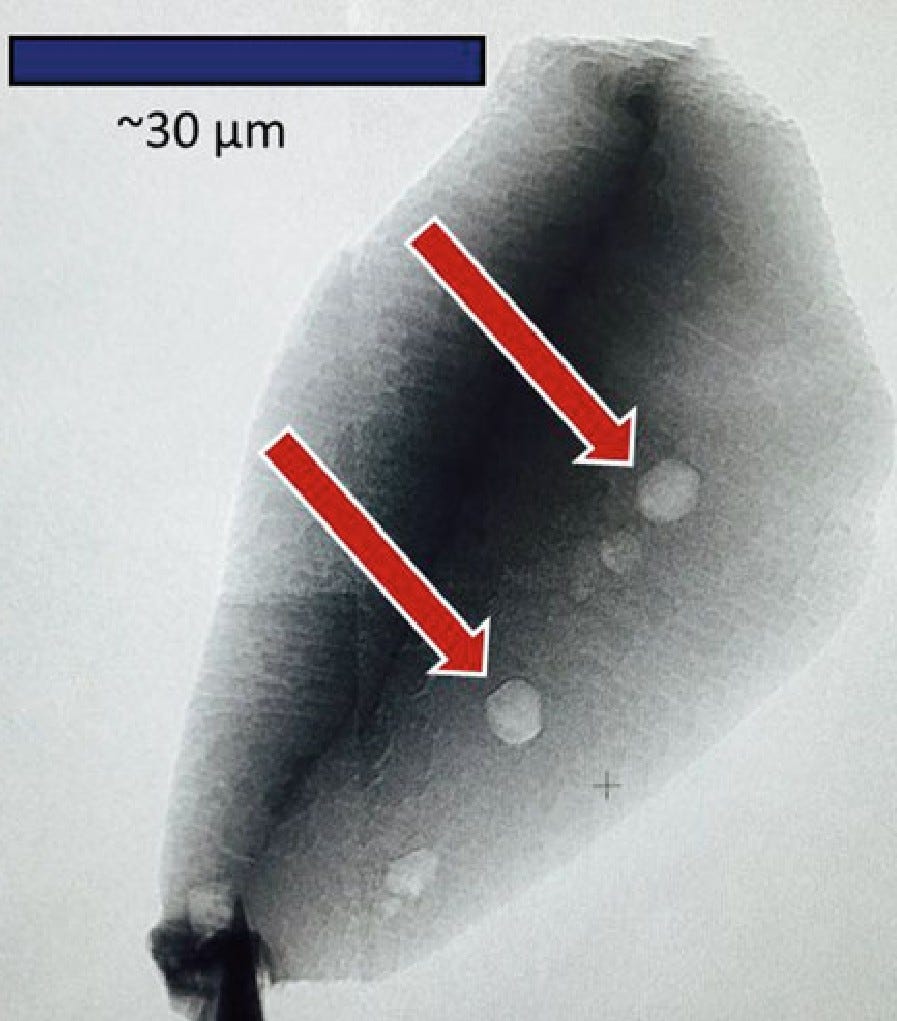
In this case, the carbon sample came from two microscopic specks of graphite embedded in a sliver of zircon, one of 566 zircon specimens from the Jack Hills of Western Australia, which are thought to contain some of the oldest minerals on Earth.
There are two striking features: (1) The sample came from 4.1 billion years ago. There was no “rock” on earth at the time. It was considered a bubbling hot cauldron. (2) There has never been any evidence before this discovery that anything like “life giving graphite” existed in the Hadean Eon, 4.5 to 4 billion years ago. This was considered impossible by geologists until this find. It was so controversial that some researchers thought it must have been contaminated during cutting the rock. But, then it was found through a Raman spectrometer that “sees through” rocks without cutting.
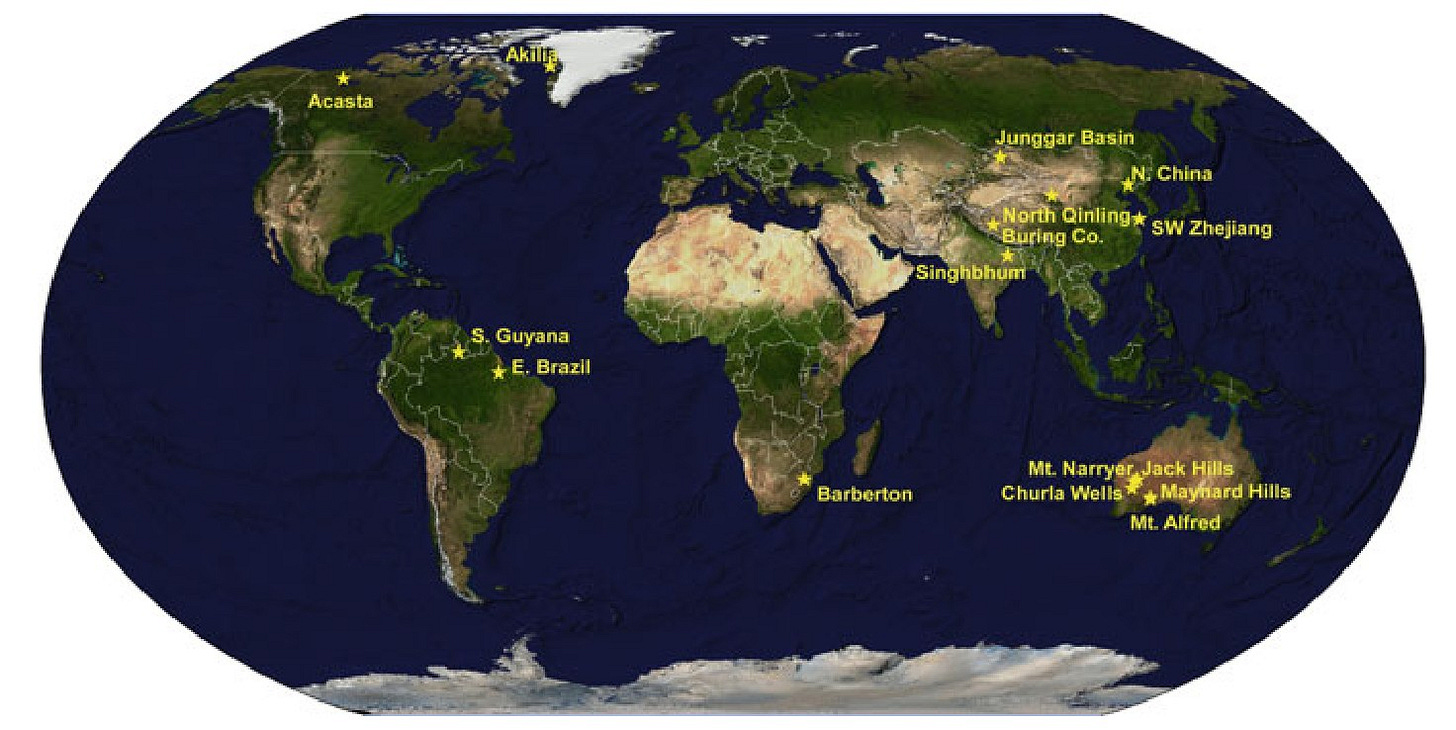
Since this discovery, fifteen locations around the globe have yielded the presence of zircon older than 4 billion years. However, no exhaustive analysis was done in any of the samples from other places to search for graphites found in Jack Hills. Why? Lack of funds.
T Mark Harrison has a rhetorical question.
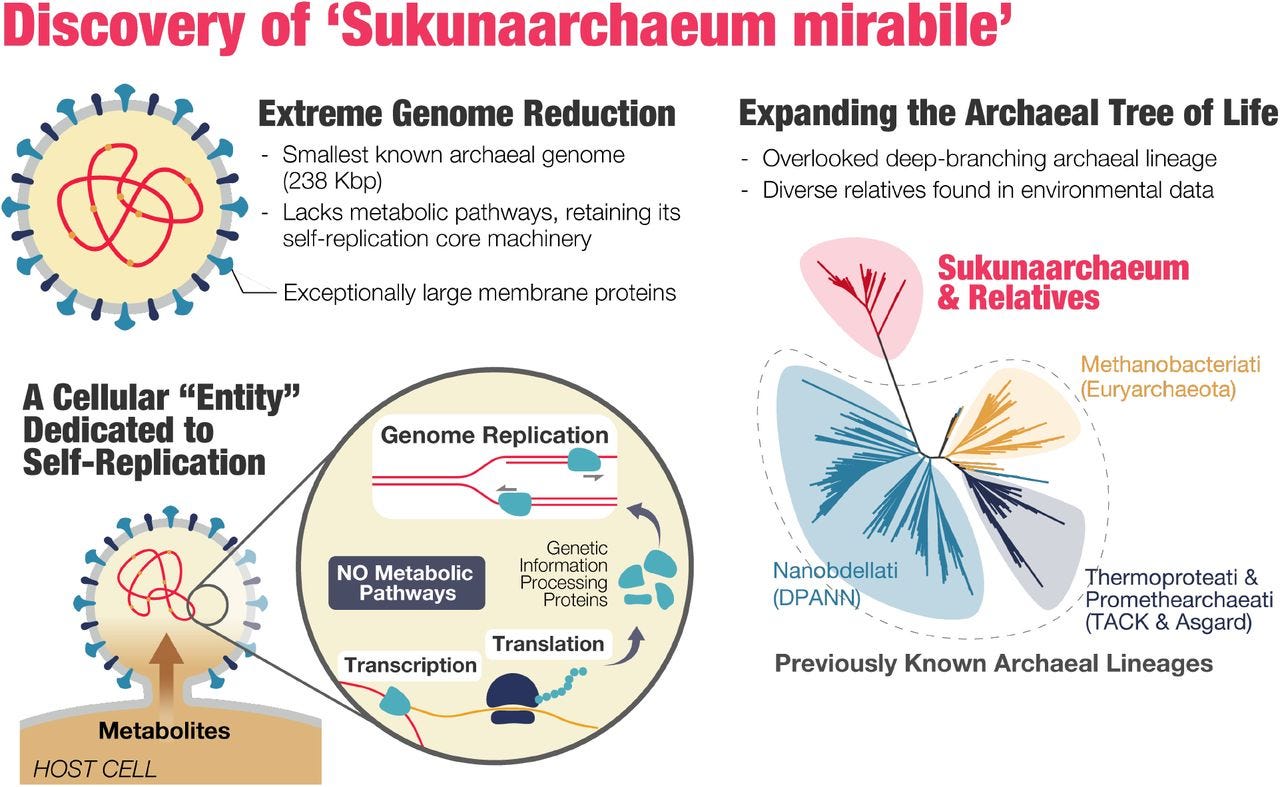
Tailpiece: A new discovery between life and nonlife Sukunaarchaeum mirabile. Like a virus, this new organism ‘Sukunaarchaeum mirabile’ outsources some functions to its host, but can still create its own ribosomes and RNA. Because virus don’t grow, reproduce on their own, or make their own energy, they’re typically excluded from definitions of life.
Its genome is also surprisingly small, and is roughly half the size (238,000 base pairs) of the next-smallest archaeal genome.
this entity contains the necessary genes to create its own ribosomes and messenger RNA—something your typical virus lacks. But like a virus, it offloads certain biological functions onto its host and it appears singularly obsessed with replicating itself.
Bottom Line: Sukunaarchaeum mirabile blurs the line between life and nonlife.